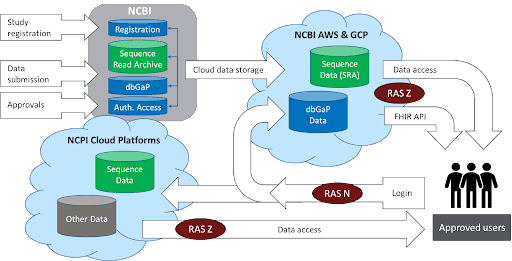
National Center for Biotechnology Information (NCBI)
NCBI hosts and manages the Database of Genotypes and Phenotypes (dbGaP) and NIH’s Sequence Read Archive (SRA). dbGaP provides and manages access to protected data related to human studies that have investigated the interaction of genotype and phenotype. In partnership with the NIH Science and Technology Research Infrastructure for Discovery, Experimentation, and Sustainability (STRIDES) Initiative, NCBI has made the entire corpus of SRA and computational tools accessible on the cloud (commercial and open access) in addition to NCBI’s local servers.
NCBI is actively developing technical solutions that modernize and streamline secure access to controlled-access data via GA4GH Data Repository Service (DRS) and NIH Researcher Authorization Service (RAS).
The central goal is to create an equitable and interoperable ecosystem where NIH-funded data is FAIR (findable, accessible, interoperable, and reusable) and NCBI is also an engaged partner in the development of community-driven solutions to provide secure access to protected data in the federated data access landscape.
Funder
This work was supported by the National Library of Medicine (NLM), NIH Office of Data Science Strategy (ODSS) and STRIDES.
PIs
NCBI
Kim Pruitt, Valerie Schneider, Kurt McDaniel, Ravinder Eskandary, Kurt Rodarmer Sr, Lon Phan, Mike Feolo, dbGaP Team, Rodney Brister, Yuriy Skripchenko, SRA team.
OER
Julia Slutsman’s team.
OSP
Taunton Paine’s team.
CIT
Jeff Erickson’s and Rebeka Rosen’s teams.
ODSS
Coordination support.
Institutions
NIH Institutes and Centers, NIH GDS Taskforce including representatives of IC Genomic Program Administrators and Data Access Committees.
Acknowledgement
We are grateful to submitting researchers for sharing their data in dbGaP and SRA, and to researchers who request access to these data to further scientific knowledge.
Datasets
dbGap Data
| Studies | 1,865 |
| Subjects | ~2.9 Million |
| Samples | ~3.4 Million |
| Phenotype: Variables | 370,825 |
| Phenotype: Values | ~2.5 Billion |
| Study Documents | 7,120 |
| Association Analyses | 7,883 |
| Genotype Assays (array) | ~2 Million |
| Genotype Assays (imputed) | 543,137 |
| Genotype Assays (seq derived) | 399,269 |
| Sequence (WGS SRA) | 178,288 |
| Sequence (WXS SRA) | 271,447 |
| Sequence (RNAseq SRA) | 86,879 |
| Epigenomic (SRA) | ~35,000 |
See dbGaP Summary Stats; numbers change daily.
SRA Data
Public Sequence Data (Number of Records)
| Data Format | Public Dataset (Hot) | Commercial (Hot) | Commercial (Cold) | Open Data Program (Hot) | Commercial (Hot) | Commercial (Cold) |
|---|---|---|---|---|---|---|
| Source | 0 | 0 | 14.2M | 0 | 0 | 14.2M |
| SRA Normalized | 775,619 | 7.5M | 5.9M | 13.4M | 825,126 | 0 |
| SRA Lite | 0 | 8.0M | 0 | 0 | 0 | 0 |
Controlled-Access Sequence Data (Number of Records)
| Data Format | Public Dataset (Hot) | Commercial (Hot) | Commercial (Cold) | Open Data Program (Hot) | Commercial (Hot) | Commercial (Cold) |
|---|---|---|---|---|---|---|
| Source | 0 | 0 | 749,915 | 0 | 0 | 749,801 |
| SRA Normalized | 0 | 608,671 | 141,244 | 0 | 705,101 | 44,700 |
| SRA Lite | 0 | 1,751 | 0 | 0 | 0 | 0 |
Services
Search
- Entrez/ SOLR indexing public metadata.
- Cross linking accessions across dbGaP, BioProject, BioSample and SRA dbs.
Request
- dbGaP Controlled Access system
- PI Selection / Reporting
- DAC Review
Data Access/Download
- RAS Clearinghouse (CLR): Performs auditing and validation of RAS passport tokens and dbGaP permissions on behalf of RAS clients.
- IDX: Performs object id exchange between SRA INSDC-style accessions and DRS ids.
- DRS: Resolves DRS ids to object location and validates access authorization via Clearinghouse.
- FHIR API: Provides access to dbGaP study level metadata.
Tools
- SRA Data Locator (SDL): supports finding data in appropriate locations
- SRA Toolkit: SRA Toolkit supports retrieval and conversion of data from into requested file format (FASTQ, ….)
- Cloud Data Delivery Service (CDDS): Service to request data in cold storage to be delivered to researchers' cloud bucket.
- ElasticBLAST: Handles large sequence-based queries. Cloud native, Alpha versions on AWS and GCP.
- BLAST+ in Docker: Cloud-based BLAST.
A Comprehensive list of NCBI Tools can be found here: All Resources - Site Guide - NCBI (nih.gov)
Authentication
- Prospective users must have NIH eRA, NIH Login or GSA login.gov account.
- RAS IDs are used as an authentication mechanism for controlled-access data and to gain access to dbGaP authorizations.
Authorization
- Authenticated users submit Data Access Requests through dbGaP Controlled Access system.
- Approvals are delivered to RAS as pre-authorizations to access data. These are encapsulated within RAS passport tokens.
- DRS approves requests for data access based upon consultation with the RAS Clearinghouse and permissions within RAS passport token.
Indexing
- Data objects are assigned permanent globally unique NCBI Accessions to allow for Access or Download across tools.
- Data cross-links are maintained across dbGaP, BioProject, BioSample and SRA.
- Datasets are identified through faceted search for public object-level metadata.
Data Models
The data in dbGaP are organized as a hierarchical structure of studies. Accessioned objects within dbGaP include studies, phenotypes (as variables and datasets), various molecular assay data SNP and Expression Array, Sequence, and Epigenomic marks, analyses, and documents
See: dbGaP Data Model
Architecture