NCI Cancer Research Data Commons (CRDC)
https://datacommons.cancer.gov/
Vision
Giving researchers a place where they can work together to access diverse data types for integrative analysis, furthering the goals of precision medicine and biomedical discoveries.
Mission
Provide access to standardized and harmonized cancer data in an expandable cloud-based infrastructure, enhancing the way data are shared to empower researchers to work in real time and with more connectivity.
Approach
To provide interoperable resources through federation, data harmonization, standards, and tools and services that can be reused across the research community and to enable enhanced data sharing.
Funder
The CRDC is funded by NCI Moonshot.
PIs
Anand Basu, Andrey Fedorov, Bill Longabaugh, Bob Grossman, Brandi Davis-Dusenbery, Brian O’Conner, David Pot, Melissa Haendel, Ron Kikinis, Sam Volchenboum, Chris Chute, Clare Bernard.
Institutions
Brigham and Women’s Hospital, Enterprise Science and Computing (ESAC), Frederick National Labs, General Dynamics Information Technology, Institute for Systems Biology, Oregon State University, Seven Bridges, The Broad Institute, University of Chicago, Johns Hopkins.
Data
Data Repositories
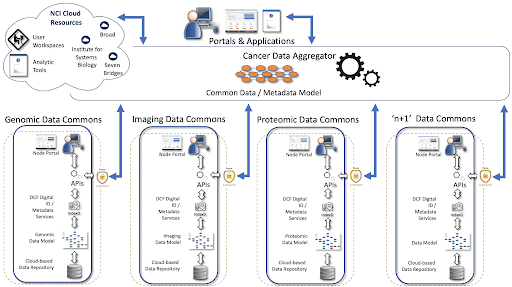
New genomic, proteomic, imaging, canine, and clinical trial data being added through both existing and new data nodes on a continual basis.
Datasets
- The Cancer Genome Atlas (TCGA)
- Therapeutically Applicable Research to Generate Effective Treatments (TARGET)
- Clinical Proteomic Tumor Analysis Consortium (CPTAC)
- Human Cancer Model Initiative (HCMI)
- Cancer Genome Characterization Initiatives (CGCI)
- Foundation Medicine (FM)
- Multiple Myeloma Research Foundation (MMRF)
- Genomics Evidence Neoplasia Information Exchange (GENIE)
- International Cancer Proteogenomic Consortium (ICPC)
- Children's Brain Tumor Tissue Consortium (CBTTC)
- Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO)
- Comparative molecular life history of spontaneous canine and human gliomas (GLIOMA01)
More Information
https://datacommons.cancer.gov/data#key-datasets
Tools
Cloud Resources
- Seven Bridges - 400+ publicly available tools and workflows in Common Workflow Language, + Dockstore, Rstudio, Jupyter notebooks, collaborative genome browser
- Broad - 700+ publicly available workflows and tools in Workflow Development Language, Integrated Genome Viewer, Dockstore, Jupyter notebooks, BigQuery, ML, pipelines
- ISB-CGC - Google: VMs, BigQuery, AI, ML, Pipelines, Cohorts, Image Viewers, Notebooks, Plotting, Dockstore
- Bring your own tools, integrative analysis is available.
Repositories Resources
- GDC: Data Analysis Visualization Exploration (DAVE) tools
- PDC: Pepquery, Morpheus, Genome Browser, DDA & DIA common data analysis pipelines
- Infrastructure: Cancer Data Aggregator (CDA), Center for Cancer Data Harmonization (CCDH), Data Commons Framework (DCF)
Analytical Tools
- List of analytical tools: https://datacommons.cancer.gov/analytical-tools
Authentication
- eRA Commons IDs (controlled data)
- NCI Data Commons Framework Services (DCFS) by Gen3
- Individual, OIDC platform authentication
Authorization
- NCI Data Commons Framework Services (DCFS) by Gen3
- Researcher Authentication Service (RAS)
- eRA Commons IDs (controlled data)
- Individual, OIDC platform authentication
Indexing
- Permanent globally unique IDs (GUIDs) for data in Google & Amazon locations
- GUIDs are cloud agnostic, promoting access and providing a mechanism for versioning data
Authorization
- dbGaP access
- DCFS by Gen3
- Authorization enabled by Trusted Partnerships with NIH
Data Models
- There are many data models across the CRDC, including ICDC, CTDC, PDC, and GDC
- Center for Cancer Data Harmonization (CCDH) develops overarching model and mapping
- CRDC also participates in GA4GH efforts
Architecture
User Perspective
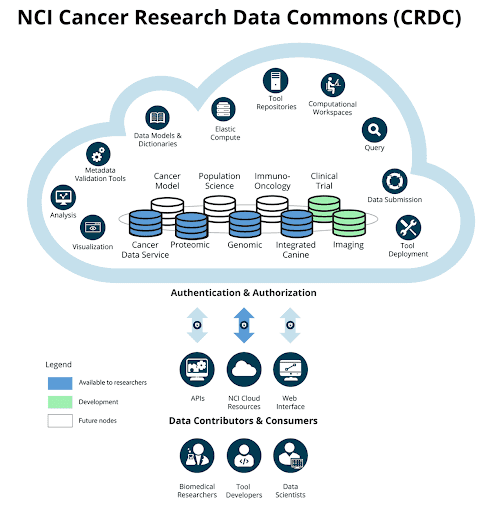
System Perspective